Unleashing Cellular Potential
Karin Schmitt, Chief Business Officer at Mogrify, explains how epigenetic switches are being used to identify the optimal culture conditions required to support the conversion and maintenance of any clinically valuable cell type

Karin Schmitt, Chief Business Officer at Mogrify
Why are epigenetic switches important for advanced/regenerative medicine?
Since Conrad Waddington first proposed the concept of an “epigenetic landscape” in 1957, researchers have worked to determine the decisive factors of cell fate. These efforts proved fruitful in the publication of Shinya Yamanaka’s discovery, “Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors,” in 2007 (1). Since then, investment in the cell therapy field has continued at pace, with many researchers and new institutes dedicated to advancing cells as therapeutics – leading to the emergence of the cell reprogramming field.
Novel approaches to cell reprogramming build on this concept by enabling the systematic identification of key regulatory switches. This may be an optimal combination of transcription factors required to drive cell identity, which make it possible to derive – via enhanced differentiation or transdifferentiation – any clinically valuable cell type.
The successful translation of reprogrammed cells into viable cell therapies, however, also relies upon the ability to culture the cells in vitro. Cell cultures need to mimic in vivo conditions as closely as possible, taking into consideration a combination of the cell’s interactions with surrounding cells, responses to environmental cues, and self-regulatory signals. Historically, cell culture conditions were determined through lengthy experimentation and trial-and-error, often based on pre-existing protocols.
Why are epigenetic switches important for advanced/regenerative medicine?
Since Conrad Waddington first proposed the concept of an “epigenetic landscape” in 1957, researchers have worked to determine the decisive factors of cell fate. These efforts proved fruitful in the publication of Shinya Yamanaka’s discovery, “Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors,” in 2007 (1). Since then, investment in the cell therapy field has continued at pace, with many researchers and new institutes dedicated to advancing cells as therapeutics – leading to the emergence of the cell reprogramming field.
Novel approaches to cell reprogramming build on this concept by enabling the systematic identification of key regulatory switches. This may be an optimal combination of transcription factors required to drive cell identity, which make it possible to derive – via enhanced differentiation or transdifferentiation – any clinically valuable cell type.
The successful translation of reprogrammed cells into viable cell therapies, however, also relies upon the ability to culture the cells in vitro. Cell cultures need to mimic in vivo conditions as closely as possible, taking into consideration a combination of the cell’s interactions with surrounding cells, responses to environmental cues, and self-regulatory signals. Historically, cell culture conditions were determined through lengthy experimentation and trial-and-error, often based on pre-existing protocols.
How can Mogrify's platform technology help identify epigenetic switches driving cell identity?
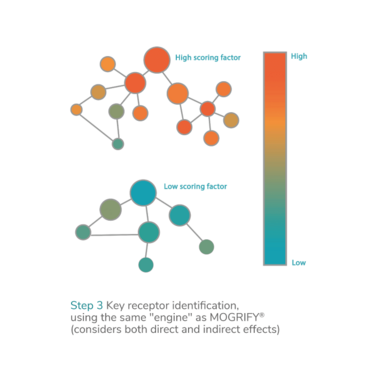
To identify optimal culture conditions, epiMOGRIFY models the epigenetic state of the cell to predict the epigenetic switches – or epigenetically predicted factors – important for the control and maintenance of cell identity. The approach is an extension of the company’s proprietary cellular conversion technology, can be applied in cGMP manufacture, and enhances directed differentiation or cell conversion to support the development of scalable off-the-shelf therapies.
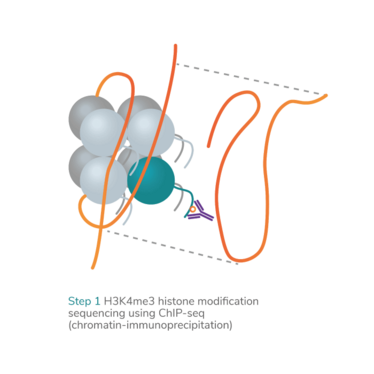
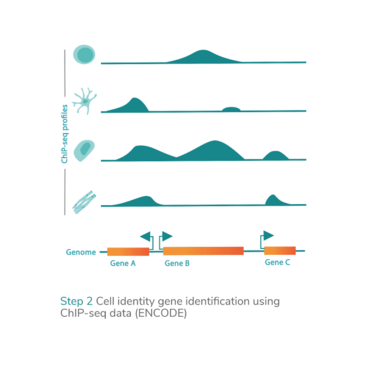
epiMOGRIFY combines gene regulatory information with a model of a cell’s epigenetic landscape and leverages changes in the level of histone methylation (H3K4me3 modifications). The platform utilizes data from more than 100 human cell/tissue types (available via the ENCODE and Epigenome Roadmap consortia) to define culture conditions that can maintain the cell identity or induce cell conversion.
How did the technology come about?
The platform, patent and knowhow were codeveloped and are co-owned by Duke-NUS (Enrico Pettreto, Associate Professor and Owen Rackham, Assistant Professor at Duke-NUS Medical School), Monash University (Jose Polo, Professor at Monash University) and Mogrify. The research underpinning this computational approach has recently been published in Cell Systems (2).
In the publication, the predictive power of the approach was validated in two ways: cell maintenance and differentiation. epiMOGRIFY-predicted factors can maintain astrocytes and cardiomyocytes in chemically defined media and promote the generation of astrocytes and cardiomyocytes from neural progenitors and embryonic stem cells, respectively. In both cell maintenance and differentiation, epiMOGRIFY-defined conditions performed as well as or better than existing undefined conditions in all cases, significantly increasing cell growth and survival and resulting in a higher differentiation efficiency.
In what ways can technologies such as these help the industry?
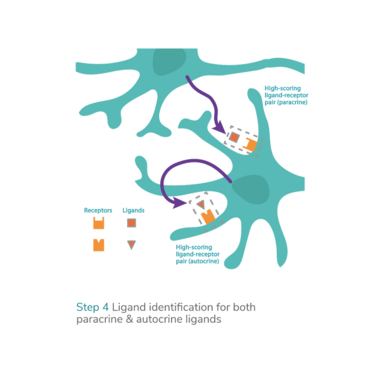
Technologies that enable the prediction and optimization of transcription factors as well as chemically defined culture medium conditions make it possible to systematically convert and maintain any target cell type from any source cell type. This addresses many of the challenges associated with the production of efficacious and scalable cell therapies: the efficient generation of clinically valuable and/or even hard-to-obtain cell types and their maintenance and expansion in chemically defined culture conditions ready for patient use. Additionally, the systemization of direct cell conversion or transdifferentiation offers new opportunities to explore novel therapeutic modalities, such as in vivo reprogramming therapies.
This article was originially published on The Medicine Maker. To access it, click the link.

Over the course of my Biomedical Sciences degree it dawned on me that my goal of becoming a scientist didn’t quite mesh with my lack of affinity for lab work. Thinking on my decision to pursue biology rather than English at age 15 – despite an aptitude for the latter – I realized that science writing was a way to combine what I loved with what I was good at.
From there I set out to gather as much freelancing experience as I could, spending 2 years developing scientific content for International Innovation, before completing an MSc in Science Communication. After gaining invaluable experience in supporting the communications efforts of CERN and IN-PART, I joined Texere – where I am focused on producing consistently engaging, cutting-edge and innovative content for our specialist audiences around the world.