Medicine Meets The Microbiome
The moment a baby enters the world – even before its first breath – the tiny body receives a massive infusion of bacteria, viruses and fungi. For thousands of years, exposure to these microbes has heralded our departure from the largely sterile environment of the womb into the real world – and all its messy complexity. No longer alone in our own bodies, we travel through life with trillions of microscopic companions. But our invisible legions aren’t simply hangers-on; studies on the body’s ecosystem have revealed the key role that the microbiome plays in health and disease. And by treating the microbiome as an extension of the human genome, we gain thousands of new targets for therapeutic intervention.
“Cut Microbe” by Rogan Brown
My interest in the human microbiome was sparked 20 years ago, when I was a fellow at Stanford University, studying immune system development and the environmental factors influencing the development of allergies and autoimmune disease. Back then, the microbiome was not the hot topic it is today. Next-generation sequencing had yet to hit the market and bacterial typing was still done by slow and laborious microbiological methods, so progress was slow.
Fast-forward 20 years, and I was working at pharma giant Pfizer, conducting research into immune-related disorders. We hoped to find mechanisms to induce the immune system to behave normally – to return it to a state of immune homeostasis – including research into gut microflora. By now, our understanding of the microbiome had come on in leaps and bounds – next-generation sequencing allowed fast and accurate sequencing of whole microbial genomes, and the human microbiome project published a reference database in 2012, mapping the normal microbiome based on 242 healthy volunteers. Big pharma can be notoriously conservative, but even they could not ignore the mounting evidence that our microbiome has a profound impact on our health. The success of fecal transplantation as a therapeutic option for treatment-resistant Clostridium difficile infection proves that the balance of gut microflora is critical for wellbeing. New research is revealing how gut bacteria impact glucose metabolism, and hence obesity and diabetes – factors behind a growing health crisis. There are also studies suggesting that the microbiome influences drug metabolism, heart health, and even our behavior (1–3). As a result, several big pharma companies - Pfizer included - have started to venture into the space, alongside smaller biotechs like Second Genome. Over $170 million has been raised in VC funding in the field to date. But it’s early days – the concepts involved are still quite foreign to companies who have been focusing on more conventional drug targets for decades.
When an opportunity came up in 2014 to head up the research program at Second Genome (www.secondgenome.com), I saw it as an opportunity to push the field forward and translate microbiomics into real clinical products. Second Genome has a well-developed bioinformatics capability to mine bacterial genomes and identify how they cause or prevent disease, and I could immediately see great potential.
A Complex Web
More than 100 trillion microorganisms live in our gut, mouth, skin and other mucosal surfaces. How do these invisible armies interact with our cells and each other? What does that mean for health and disease? We’re only just beginning to scratch (and sniff) the surface…
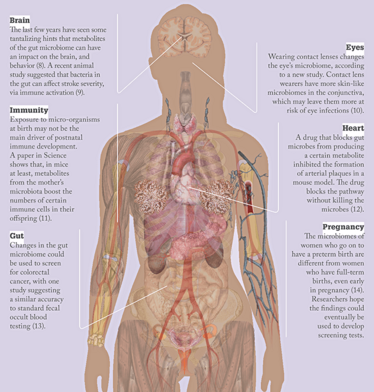
Microbes to molecules
Probiotics have an image problem – often sold as supplements in health food stores or online, their association with alternative therapies has done nothing to encourage scientists to explore the therapeutic potential of the microbiome. These are highly sensitive organisms, which depend heavily on environmental factors and the correct energy sources to survive and function effectively. Manufacturing them to a standard that ensures safety and efficacy is not a trivial task. The pharmaceutical industry and the regulatory agencies have spent the last 100 years figuring out how to deliver safe and effective small molecules and biologics – dealing with living entities that can transform and adapt in response to environmental factors is another level of complexity. The health food and supplement industry do not operate under those controls, so it’s hardly surprising if their products show little efficacy, especially in well-controlled clinical studies. Add in a few months of sitting on a shelf with no quality control and you can be sure that a large proportion of the bacteria or yeast in your daily probiotic pill are dead.
However, if you delve into the literature studying probiotics in animal models, where large doses are administered in animals with a standardized gastrointestinal microbiome, probiotics work well. There are positive examples in humans too – a small subset of post-surgical inflammatory bowel disease (IBD) patients suffering from pouchitis respond very well to a probiotic mixture (4). Nevertheless, all in all, pharma companies and clinicians remain unconvinced by the concept of live bacterial therapeutics outside of the C. difficile area.
Second Genome is taking a different approach. It was founded in 2010 by academic Corey Goodman, together with our head of bioinformatics Todd DeSantis. The company originally used Lawrence Berkley National Laboratory technology to type bacteria in environmental samples, to gain a better understanding of the bacterial ecosystems present in water, food and various other environmental samples. By 2011, the data around the human microbiome was starting to explode, and the company decided to re-orientate itself towards human microbiome analysis – analyzing human samples for the pharmaceutical, medical, and academic sectors. In 2012, the company was ready to take the next step – in addition to offering human sample typing as a service, the company launched a program to study bacterial functions in human diseases. I joined shortly after that, to help the company identify therapeutics from bacteria.
Rather than developing live bacteria as therapies, we mine the genomes of probiotic bacteria for small-molecule metabolites, proteins and peptides that mediate the putative benefits of probiotics. We start with the hypothesis that the bacteria within the human body have a biological function, and that those functions must be mediated by signaling molecules. Using human cell-based assays we intercept the conversation between our bodies and our bacteria. The aim is to produce a drug based on pharmacological effect, produced to a quality standard and predictability that meets the needs of a medicine rather than a food supplement. For every one human gene, there are 100 more genes within our microbiome – at least some of those are likely to be druggable targets.
A gut feeling
The decision to start our therapeutic research program in IBD was pretty straightforward. Firstly, a tremendous amount of clinical data has been generated over the past 15 years on the role of intestinal bacteria in IBD, and probiotics have already been found to be an effective treatment for certain patient groups. Secondly, IBD provides a perfect opportunity to connect bacterial and host responses within the same sample – exactly what our technology is designed to do. In IBD, mucosal biopsy samples are used for diagnosis, categorization, and evaluation of treatment response. Those samples offer a rare opportunity to analyze gene expression in microbes and host, in a clinical setting. Our platform also allows us to pull in primary data from other published studies and so increase our bioinformatics capability.
We’re using this approach to target bacterially induced inflammation in the gastrointestinal tract of IBD patients. We compared ulcerative colitis patient samples to those of healthy individuals, as well as those from patients who were in remission after responding well to drug therapy. After analyzing all of our data (and adding data from other studies) we found that pathways linked to activation of innate immune responses in response to bacteria were dramatically upregulated in IBD patients with active disease. On analyzing the bacteria, we found that molecules activating these innate immune pathways (for example, toll-like receptors) were also dramatically upregulated. So we chose to develop a small-molecule drug targeting bacterially induced innate immune activation. That drug – SGM-1019 – has been evaluated in a Phase I clinical trial and we’re hoping to start a Phase II proof-of-concept study in IBD patients at the start of 2017.
Can microbes make you fat?
A number of studies have been published showing that gastric bypass induces a very rapid change in insulin sensitivity for obese and diabetic patients, and that this correlated to changes in their microbiome. Jeffrey Gordon’s very elegant research looked at twins where one was of a healthy weight and the other was obese. The study found that, when their microbiota were transferred to mice raised in a microbe-free environment, the mice tended to be fat or thin accordingly (5). Of course, diet is very important too, and the evidence suggests that diet and microbiome are tightly linked. When mice were exposed to microbiomes from both fat and thin twins, diet determined which bacteria predominated – a high-fat diet with few vegetables prevented proliferation of bacteria associated with leanness.
A study published in late 2015 by researchers in Israel (6), analyzed BMI, diet, microbiome, and various other metrics for 800 people. Using machine-learning algorithms, they were able to identify how our microbiome and a few other physiological factors determine our response to a specific diet; essentially, our microflora composition changes the amount of glucose that is extracted from our food. Microbes metabolize all sorts of nutrients and transform them into bioactive molecules. In fact, an estimated one-third of circulating metabolites are a product of the gut microbiota.
Obesity is the result of a complex mixture of diet, individual behavior, drugs, and so on – these factors interact to influence not only the microbial composition, but the metabolic activity of those bacteria in our gastrointestinal tracts. The metabolites of the gut bacteria in turn affect liver cell behavior, influencing insulin sensitivity and the amount of energy that we extract from our diet. We’re currently carrying out studies to analyze the metabolic machinery of intestinal bacteria in obese patients and detect the resulting metabolites in the blood. Specifically, we’re looking at patients undergoing gastric bypass to find out how a restricted diet and dramatically altered microbiota correlate to physiological changes. We want to understand how the types of bacteria are changing, how their pathways are changing and how they are influencing host responses when it comes to insulin resistance, and so on.
At first glance, our two disease foci – IBD and metabolic disorders – seem disparate, but the underlying mechanisms overlap. Inflammation appears to be a common link – low-grade in metabolic disease, much greater in IBD. In addition, we believe diet, host genetics, environment and microbiota play a part in both areas – they are both abnormal physiological responses to diet and the environment.
Rogan Brown
The law of unintended consequences
Given the sheer complexity of host–microbe and microbe–microbe interactions, there is of course a risk that changing the microbiome could have unforeseen consequences. Until we get to a point where we have a complete systems biology understanding of how alterations in one area will influence the entire system, there is always a risk. I recall reading an anecdotal report about a previously healthy weight C. difficile patient in the UK who received a fecal transplant from her overweight daughter – the procedure cleared the patient’s infection but she gained weight rapidly and became obese, leading some doctors to avoid overweight donors for fecal transplant (7). Accidental effects on the microbiome are a significant concern, and one reason we have chosen to focus on very specific signaling molecules rather than live bacterial therapy or probiotics. We want to move the field towards mechanistic, science-based drug discovery, in line with any other pharmaceutical drug.
Unintended changes to the microbiome are not limited to our attempts at manipulation. For example, the widespread use of antibiotics not only increases antimicrobial resistance but also has an undetermined effect on the microbiome. Such changes are not always easily apparent in two or six week studies, and may sometimes only manifest themselves over the course of a lifetime. This realization is particularly worrying when we look at antibiotic use in early life and consider that it could have consequences in middle age. Cesarean section could also affect a newborn baby’s early microbiome by dramatically delaying microbial colonization (read more below about how medical interventions in early life affect the microbiome in “The Changing Microbiome”).
Clinical connections
I believe that we are moving towards an understanding of human physiology that integrates microbial biology – the microbiome will be seen as an extension of the genome. In the future, we will no longer consider human biochemical pathways in isolation, but instead adopt a much more extensive integration of microbial biology into our understanding of mammalian biology. From there, I think we will begin to use manufactured live bacteria as therapies, potentially engineered to boost their effectiveness.
Ultimately, once we fully understand the complex biochemical interplay between microbes and mammalian systems, we will have opened a new gateway into improved health for all.
Karim Dabbagh is Chief Scientific Officer at Second Genome, Inc, San Francisco, CA, USA.
The Changing Microbiome
Martin Blaser has been studying the complex relationship between microbes and the human body for over 30 years. Known for his groundbreaking work on host–bacterial interactions in Helicobacter pylori infection, in recent years he has called attention to medical advances that could permanently disrupt the human microbiome. We caught up with Blaser to find out more.

How did you first get interested in the microbiome?
I’m a medical doctor and my specialty is infectious diseases. I began studying pathogens in 1977, in particular H. pylori in the stomach. Initially, my work focused on eliminating H. pylori, which can cause stomach ulcers, but later I discovered that it might also have positive effects in the body. That got me thinking about the duality of organisms that live in our bodies – they can harm us, but also help us. From there, I began to focus more and more on how organisms can help us.
The human microbiome is a scientific frontier that has really emerged in the last decade, and continues to advance rapidly. We have the general outlines of many of the important processes, but we are missing many of the details.
Tell us about the human microbiome project (HMP)
The HMP was launched by the National Institute of Health (NIH) in the United States to advance our knowledge of the human microbiome. It involved a large project examining the microbiome of 250 healthy volunteers, and demonstration projects looking at the relationship between the microbiome and different diseases, such as psoriasis, inflammatory bowel disease and obesity. It was an expensive project, costing the NIH about $130,000,000, but it taught us a lot, and formed the foundation for many of the studies that are going on today.
Were there any surprises from the HMP?
One of the biggest surprises was how the species of bacteria are so varied across populations – different people have different bacteria carrying out the same functions. A lot of our microbiome is inherited from our parents, specifically from our mothers during birth.
How is the microbiome being changed?
Being exposed to the bacteria in the mother’s birth canal is a big part of how babies populate their microbiome in early life – cesarean section removes that opportunity. Cesarean sections can be lifesaving in a small percentage of cases, but in the US we’re up to 32 percent of babies being born by cesarean section and in some countries we’re above 50 percent, so it seems clear it is being overused. I’m also concerned by how often young children and pregnant mothers are prescribed antibiotics.
Mothers have been passing on microbes to their babies for millions of years. Antibiotics, cesarean sections, and all kinds of antimicrobial cleaners are changing that normal transmission. That could have serious consequences down the line. We have to start thinking about the microbiome when weighing up the costs of antibiotic use or cesarean sections.
- TA Clayton et al., “Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism”, Proc Natl Acad Sci USA 106(34), 14728–14733 (2009).
- Z Wang et al., “Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis”, Cell 163, 1585–1595 (2015).
- P Bercik et al., “The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice”, Gastroenterology 141, 599–609 (2011).
- T Kühbacher et al., “Bacterial and fungal microbiota in relation to probiotic therapy (VSL#3) in pouchitis”, Gut 55, 833–841 (2006).
- VK Ridaura et al., “Cultured gut microbiota from twins discordant for obesity modulate adiposity and metabolic phenotypes in mice”, Science 341, 1241214 (2013).
- D Zeevi et al, “Personalized nutrition by prediction of glycemic responses”, Cell 163, 1079 (2015).
- N Alang, CR Kelly, “Weight gain after fecal microbiota transplantation”, Open Forum Infectious Diseases, 2, ofv004(2015).
- Y Wang, LH Kasper, “The role of microbiome in central nervous system disorders”, Brain Behav Immun 38, 1–12 (2014).
- C Benakis et al., “Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells”, Nature Medicine,PMID: 27019327 (2016).
- H Shin et al., “Changes in the eye microbiota associated with contact lens wearing”, mBio 7, e00198–16 (2016).
- M Gomez de Agüero et al., “The maternal microbiota drives early postnatal innate immune development”, Science 351, 1296–1302 (2016).
- Z Wang et al., “Non-lethal Inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis”, Cell 7, 1585–1595 (2015).
- G Zeller et al., “Potential of fecal microbiota for early-stage detection of colorectal cancer”, Mol Syst Biol 10, 766 (2014).
- DB DiGiulio et al., “Temporal and spatial variation of the human microbiota during pregnancy”, Proc Natl Acad Sci USA 112, 11060–11065 (2015).

Karim Dabbagh leads the R&D organization at Second Genome. Prior to that, he led the immunoregulation department at Pfizer, an R&D group focused on innovative approaches to elicit homeostatic immune responses, including microbiome research, for the treatment of immune related disorders. At Pfizer, he also led external R&D innovation for autoimmune and inflammatory diseases. He grew up surrounded by science fiction, and believes one day we will be able to harness the microbiome for nutritional and therapeutic benefits.