
Viruses Face the Ultimate Test
Shotgun sequencing is a powerful tool in microbial genomics, but viral genomes have proven tough to crack. Enter ViroCap and its ability to enhance shotgun sequencing to detect the vast majority of viruses infecting humans and animals in a single test. For viruses, is there anywhere left to hide?
Ever since Watson, Crick, Wilkins and Franklin's discovery of the double helix, we have longed to unlock the secrets of every base pair. The completion of the Human Genome Project, while an amazing achievement, was just the start – we immediately set to work on mapping as many variations, mutations and interactions that influence our genes in health and disease as we could. And why stop at the human genome? After all, the very first genomes to be sequenced were those of microbes, and there are obvious benefits to knowing the genetic makeup of the thousands of microorganisms that live within and alongside us, especially those that cause disease. The development of new, faster genome sequencing technologies has made it feasible to sequence any microbial DNA found in a sample – the basis of the relatively new science of metagenomics.
(Shot)gunning for viral genomes
It was the identification of a gene back in 1971 – 16S ribosomal RNA – found in all bacteria, that made molecular identification of bacterial pathogens relatively simple, explains Greg Storch, Professor of Pediatrics at Washington University School of Medicine. Greg, who assisted Kristine and Todd Wylie with the development of ViroCap says, “There is no single gene that we can amplify across all viruses, and that’s why we have to take the much broader approach of metagenomic shotgun sequencing (MSS).”
MSS, in which all the DNA – and sometimes RNA – in a sample are (i) fragmented at random, (ii) sequenced, and (iii) classified based on comparison to known reference genome sequences, has been a useful tool for studies of the microbiome. Kristine Wylie, Assistant Professor in the Department of Pediatrics at Washington University School of Medicine, has previously used MSS both to examine the normal human virome as part of the Human Microbiome Project, and to detect the presence of viruses in children with unexplained fevers. “We loved the approach of sequencing because we were able to look at all viruses without having any preconceptions of what we might be looking for, or what we might find,” she says. However, MSS is limited by the small size and extensive variability of many viral genomes, which leads to poor sensitivity. Kristine and her collaborators thought they could do better.
“The initial idea came from work we had previously done with whole-exome sequencing,” explains Todd Wylie, Instructor in Pediatrics at Washington University School of Medicine. Exome sequencing uses targeted capture to sequence only genes, which is quicker and cheaper than whole-genome sequencing because noncoding sequences account for the majority of the human genome. “We thought that, using the same idea, we could build a panel capable of targetting other species, such as bacteria or viruses, to enhance the sensitivity of MSS,” says Todd. Focusing their efforts on viruses, the team created “a comprehensive viral targeted sequence capture panel that could be used to assess all viruses known to infect vertebrate cells and detect divergent viruses” – or, more concisely, ViroCap (1).
If the ViroCap fits...
ViroCap is an MSS enhancement tool that provides the sensitivity required to accurately detect virtually all previously sequenced genomes of viruses infecting vertebrates, plus viruses that have yet to be sequenced but share some of their genetic code with a virus on the panel. The test targets 34 viral families, with 190 annotated genera and 337 different species, shown in Figure 1. A comparison of ViroCap viral sequence reads versus standard MSS can be seen in Figure 2. The results are impressive, and the new tool has understandably generated a lot of excitement.
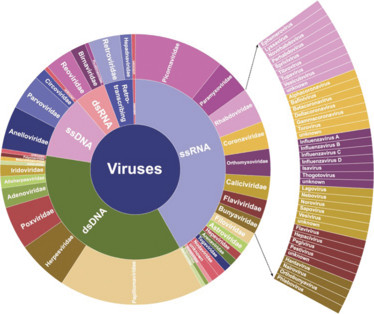
Figure 1. Taxonomic distribution of target genomes included in ViroCap (1).
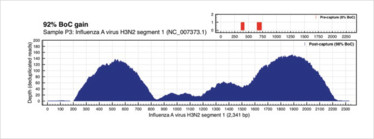
Figure 2. Targeted sequence capture enrichment, influenza A virus (1).
“In relation to detecting viruses that are actually present in the sample, it appears to be very powerful because we’re able to obtain a very large proportion – up to 100 percent – of the sequence of viruses that are in clinical samples. Once we have that much sequence, there’s very little doubt about the identity of the virus”, says Greg. Even a group of viruses with a very high degree of variability – the anelloviruses – were identifiable by ViroCap.
Several microarray-based techniques are available for viral characterization and discovery; however, they only target relatively small, discrete regions of the viral genome, while ViroCap targets the complete genome. As a result, microarrays can only indicate that the virus is present, whereas ViroCap provides sequences – and sometimes the complete genome. By providing instant information on taxonomy, strain typing, virulence characteristics, and anti-viral drug resistance genotype, ViroCap has the potential to be a game-changing technology for both research and clinical applications.
Translation will take time
A test able to detect and identify almost any viral pathogen has obvious applications for clinical diagnostics. However, it’s likely to take years before it reaches doctors, according to Todd: “To get ViroCap into the clinic, as with most new inventions, we will need to increase the speed and decrease costs.”
Greg agrees, “It’s relatively slow and expensive in its current form so you can think of the work we have done so far as a proof-of-concept for clinical laboratory testing. But we anticipate that rapid advances in technology and refinements to the technique will lead to progress in both speed and cost. We do think that this will come into clinical testing over the next few years.”
Initial results hint at the potential diagnostic power of the technique – in a study of children with unexplained fevers, ViroCap increased the breadth of coverage from 2 to 83 percent compared with MSS, and identified several viruses not picked up by MSS at all.
Those looking to harness ViroCap to analyze clinical samples will need to bear in mind that the results won’t provide information on clinical significance. “ViroCap is not judgmental. It reveals the presence of viruses in a clinical sample, but additional information would be required to determine whether the virus or viruses are significant,” says Greg, “You definitely need a human interrogator to interpret the information.”
The novelty of the approach could also bring up regulatory challenges. “It’s not clear what the path would be to FDA approval of a technology like this, where the capability of detection is very open. It’s an area that’s under very active discussion, both in microbial genomics and human genomics. The big question is what the FDA and other regulatory agencies will require for tests based on next-generation sequencing,” says Greg.
Research ready
For now, the team is focusing on how ViroCap can be used in microbiology and infectious disease research. “We are very interested in applying this technology to a whole variety of questions – both clinically relevant and more fundamental research questions,” says Greg. “In particular, we want to apply it to diseases of unknown etiology, where there’s some reason to be suspicious that viruses are important. One of the big areas is neurologic disease, such as encephalitis or meningitis, and another area is fever of unknown origin, both in adults and children, where current diagnostic testing does not always reveal an answer.”
The response to the technology has been overwhelmingly positive, say the researchers. “It’s generated a lot of interest and excitement in our field, and in the scientific community and the public at large,” says Todd.
Kristine says that scientists doing similar work immediately saw the potential for the technique. “There are a lot of scientists trying to use sequencing to look at viruses, and we’ve all experienced the same problems with sensitivity. So people are very excited for their own research, because this solved a problem that they have all been running up against.”
Collaboration goes viral
The team has already been approached by researchers working in a wide variety of research areas, according to Greg: “ViroCap has very broad-based applications because it has the potential to enhance the detection of all viruses that infect vertebrates. So we’ve heard from a very diverse selection of people in a variety of settings.”
The team is making the technology available to researchers immediately. The reason is simple, says Greg, “We are scientists, and we want to promote the scientific process. ViroCap is very big, and we think that researchers around the globe will find applications for it, and we want the scientific process to move forward.”
“When Washington University was involved in the Human Genome Project they made the sequences determined each day available that night. We are continuing that tradition,” Greg says. Researchers who would like to make use of the technology are encouraged to contact the researchers directly.
Looking forward, the team plans to keep developing their informatics pipeline, while working on decreasing the time and cost of ViroCap. They also hope to overcome the limitations that may be holding ViroCap from its full potential, including filtering out bacterial vector sequences and building up the viral library to give the largest possible range of detectable sequences. The team even suggests that ViroCap could be modified to become a tool for broader pathogenic identification, including the detection of other human pathogens, such as bacteria, fungi and protozoa – a truly universal test.
“There is no end to the potential applications”, concludes Greg.
My fascination with science, gaming, and writing led to my studying biology at university, while simultaneously working as an online games journalist. After university, I travelled across Europe, working on a novel and developing a game, before finding my way to Texere. As Associate Editor, I’m evolving my loves of science and writing, while continuing to pursue my passion for gaming and creative writing in a personal capacity.















