Small(er) Sequence, Big(ger) Promise
Efficient CRISPR gene editing with a newly characterized CjCas9 orthologue might bring the approach one step closer to the clinic
At a Glance
- CRISPR/Cas9 gene editing shows much promise therapeutically, and since its debut, there’s been lots of research into developing and using the technique
- But although SpCas9 and SaCas9 are currently the most commonly used Cas9 orthologues for CRISPR/Cas9 gene editing, each has limitations for gene therapy approaches
- According to recently published research, a team from South Korea might have found a promising alternative – CjCas9
- SeeokJong Kim and Sung Wook Park, co-authors on the paper, tell us about their work and what’s next
There is little doubt that the future of medicine is gene editing. But right now, the focus is on figuring out how to get there as safely and effectively as possible. An approach at the forefront of our gene-editing endeavors is CRISPR (clustered regularly interspaced short palindromic repeats)/Cas – and since its debut in 2013 (1)(2)(3), research has been booming.
Although CRISPR/Cas9 shows much promise therapeutically, improvements and tailored modifications are needed before it hits the clinic. For example, the Cas9 endonuclease – which binds and cuts DNA at specific locations as dictated by a short guiding RNA (sgRNA) sequence (Figure 1) – could benefit from optimization; the most commonly used orthologue (derived from Streptococcus pyogenes – SpCas9) weighs in at a mighty 4.10 kbp and 1,368 amino acids, and is too big to be packaged into a single adeno-associated virus (AAV) vector along with its sgRNA sequence. It can be split over more than one AAV vector, but this can come at the cost of reduced endonuclease activity (4). One alternative is Cas9 from Staphylococcus aureus (SaCas9); it’s around 1 kbp smaller than SpCas9 and can be packaged into a single AAV vector. Unfortunately, the number of targetable genes is predicted to be limited by a much less frequently occurring protospacer-adjacent motif (PAM) – a 2–6 base pair DNA sequence that acts an essential targeting component of the system.
Given the apparent lack of “good” choice, a team in South Korea decided to seek alternative Cas9 orthologues – with promising results. In their recently published study (5) they characterized Cas9 from Campylobacter jejuni (CjCas9), demonstrated its use for in vitro and in vivo gene editing, and showed that CjCas9 targeted to Vegfa or Hif1a could reduce choroidal neovascularization (CNV) in a mouse model of age-related macular degeneration (AMD; See Box – Summary of Key Results). Seokjoong Kim, Research Director of Toolgen, and Sung Wook Park, one of the lead authors on the paper, tell us more.
What inspired your study?
Seokjoong Kim: I’m a molecular biologist, and over the last 10 years I’ve been working to develop gene editing tools such as transcription activator-like effector nucleases (TALENs) and CRISPR. When we were ready to move into the clinical translation of CRISPR technology, I found AAV very attractive because it has been clinically proven to deliver genes very efficiently and safely. I wanted to combine a CRISPR/Cas9 system with AAV, but I quickly found out that the typical Cas9 system from Streptococcus pyogenes is simply too big. We started to use Cas9 systems from different species, but decided to focus on CjCas9 because it was the smallest we could find in the literature and databases. In collaboration with The Institute for Basic Science in Seoul, we then performed a full characterization of CjCas9, including its PAM sequence and the optimal size of the sgRNA. We showed that we were able to support efficient gene editing in vitro and in vivo with CjCas9. Our initial trial was in muscle, but we later focused on gene editing in the eye to establish a therapeutic effect. We chose the eye because it is an isolated organ and easy to access, which resulted in our collaboration with Jeong Hun Kim and Sung Wook Park, and the focus on AMD and diabetic retinopathy (DR).
Sung Wook Park: We’d been looking for an efficient tool for genome editing in the eye. Because Cas9 is too large to package into AAV vectors without “splitting up,” we were trying to find different ways to pack it into AAV vectors. Biotech company Toolgen had already begun developing CjCas9, so we got involved.
What are your most important findings?
SK: We are pleased that we have been able to develop the Campylobacter jejuni CRISPR-Cas9 system and show that AAV-mediated delivery can support very efficient gene editing in the eye – we saw editing efficiencies of 30–60 percent. Fortunately, the dose efficiency of gene editing was enough to show some phenotypic changes, and we were happy to see that targeting Vegfa and Hif1a in the eye with CRISPR gene editing could change the phenotype in a mouse model of laser-induced CNV.
SWP: The delivery method is really quite different. Most gene therapy relies on AAV2, which is typically delivered through subretinal injection, which inevitably causes damage to the retinal layer. We already had experience in using AAV9 for targeting retinal pigment epithelium (RPE) cells, and we delivered the CjCas9 AAV9 viral vectors via intravitreal injection. We believe intravitreal injection is a much better delivery method because we limit damage to the retinal neurons. Also, simply injecting the viral vector into the vitreal cavity can increase efficiency because more cells in the retina or RPE can be transduced and edited.
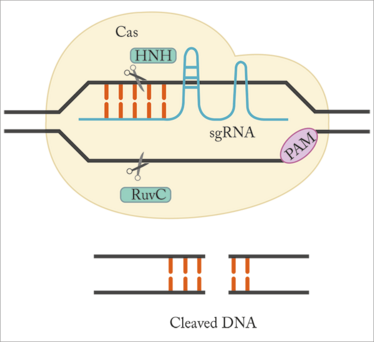
Figure 1. CRISPR gene editing. Cas9 binds to DNA guided by the sgRNA sequence, and the HNH and RuvC nuclease domains of Cas9 cleave the DNA. For Cas9 to bind and cut the DNA, a PAM sequence must be present immediately downstream of the target sequence recognized by the sgRNA. Adapted from (6).
Summary of Key Results
- At 2.95 kbp and 984 amino acid residues, CjCas9 is around 30 percent smaller than SpCas9
- Cleaving of human genomic DNA in vitro by CjCas9 was found to be more specific than SaCas9 with no reduction in efficiency
- In vivo, CjCas9 was shown to induce targeted mutations in three genes: – In mouse myotubes, CjCas9 induced targeted mutations at the Rosa26 locus. – In mouse retina, targeted mutations were induced in Rosa26, Vegfa and Hif1a in retinal pigment epithelium (RPE) cells
- In a laser induced CNV mouse model, the team found that targeting Vegfa and Hif1a each reduced the relative CNV area by over 20 percent (see Figure 2). Hif1a encodes hypoxia-inducible factor alpha (HIF-1α), a transcription factor that activates the transcription of VEGF-A.
- To investigate the potential side effects of the partial knockouts of Vegfa and Hif1a in RPE cells, cone function was measured by full-field electroretinography. Photopic and flicker response were not significantly decreased, but the size of the opsin-positive area was reduced when Vegfa was targeted.
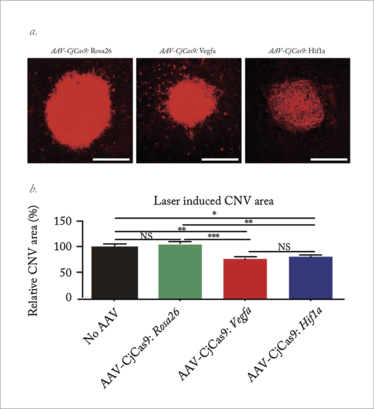
Mice injected with AAV-CjCas9 targeted to Rosa26, Vegfa or Hif1a were treated at day 42 post-injection to induce CNV; one week later they were analyzed. a. Representative images showing the area of laser-induced CNV (stained with isolectin B4). Scale bar, 200 mm. b. Graph showing the relative CNV area. Error bars indicate SEM. (n=17–18). One-way ANOVA and Tukey’s post hoc tests, *P<0.05; **P<0.01; ***P<0.001; NS, not significant; SEM, standard error of the mean. Adapted from (5).
Any surprises or challenges along the way?
SK: When I initially chose the study, I thought that the production of AAV would be well established because there had been so many trials – so I was surprised at how inefficient and expensive it is, even at the research level! With the ongoing work on AAV-mediated gene therapy in the US and Europe, I hope that in the near future we will have more established and optimized protocols and processes for AAV production.
SWP: It isn’t proven in well-established studies, but there is some evidence that targeting the Vegfa gene can cause problems, so we needed to check the side effects. There was some local opsin decrease but there were no constant functional decreases in photoreceptor response. So at the present time, we believe that targeting Vegfa is still a viable option.
When do you expect to move into human trials?
SK: It’s hard to say! We believe we can perform some large animal studies within this year – and we hope that we’ll be able to prepare enough data to file an investigational new drug (IND) application in 2018. One thing for us to consider is the fact that we are currently targeting Vegfa and Hif1a –but these are not actually defective genes in AMD and DR. We do believe that targeting these genes – and others that we are working on – could be a viable option for long-term AMD or DR therapy, but we’re not sure if regulatory bodies will be comfortable with targeting non-defective genes with CRISPR. Therefore, we are also interested in targeting other genetic diseases in the eye that might be more easily accepted by regulatory bodies and society.
SWP: We also have to consider the hurdles to overcome before getting to trials. As well as the necessary larger animal and safety trials, we need to find an effective human sequence and prove that it can be edited with the CRISPR-CjCas9 system. Hif1a is quite conserved between the mouse and human genomes, so we are thinking of targeting this in clinical trials. Additionally, because anti-VEGF therapy is a well-established treatment regimen for AMD and DR, we’ll need to demonstrate how effective CRISPR is compared with existing therapies. These diseases usually wax and wane over time, so multiple repeat injections of anti-VEGF agents are needed; we hope that our genome editing approach might be able to downregulate VEGF and other factors to below the threshold level that causes disease.
Where do you hope your work will take you?
SWP: Using the advantage of the small size of CjCas9, we might be able to think about combinational therapy approaches where we target dual genes with a single AAV system. We haven’t tried this yet, but we are looking into the possibility of different approaches.
SK: There is no limit to the genes you can edit with the CRISPR-Cas9 system. VEGF is a key target for modulating vascularization, firstly because it is one of the very important factors in the process, but also because it can be inhibited extracellularly by current therapy models like monoclonal antibodies. We’re also looking at genes for intracellular or nuclear proteins that cannot be easily modulated by antibodies or small molecules. Looking forward, we want our technology to bring new hope to patients who are losing sight.
Seokjoong Kim is Research Director of ToolGen, Seoul, Republic of Korea.
Sung Wook Park is a Researcher at Department of Biomedical Sciences, Seoul National University College of Medicine and FARB Laboratory, Biomedical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea.
- L Cong ET al., “Multiplex genome engineering using CRISPR/Cas systems”, Science, 339, 819–823 (2013). PMID: 23287718.
- P Mali et al., “RNA-guided human genome engineering via Cas9”, Science, 339, 823–826 (2013). PMID: 23287722.
- M Jinek et al., “RNA-programmed genome editing in human cells”, Elife, e00471 (2013). PMID: 23386978.
- B Zetsche et al., “A split-Cas9 architecture for inducible genome editing and transcription modulation”, Nat Biotechnol, 33, 139–142 (2015). PMID: 25643054.
- E Kim et al., “In vivo genome editing with a small cas9 orthologue derived from Campylobacter jejuni”, Nature Communications, 8, 14500 (2017). PMID: 28220790.
- EJ Kim et al., “CRISPR-Cas9: a promising tool for gene editing on induced pluripotent stem cells”, Korean J Intern Medicine, 32, 42–61 (2017). PMID: 28049282.
Following my journey through academia, I entered the world of scientific writing and never looked back. After several years of working as a medical writer – where I developed a wide range of skills in healthcare publications and communications – I took the opportunity to stretch my creative and journalistic muscles and joined Texere Publishing. Working as Editor on The Ophthalmologist allows me to nurture my skills in scientific writing – and explore the dynamic world of ophthalmology – all within an innovative and exciting company.















