
MSI Marvel
Eva Cuypers discusses the almost endless potential of mass spectrometry imaging (MSI) in clinical applications, her research on intraoperative diagnostics for the rapid identification of cancer cells, and the importance of researcher–clinician relationships
Margot Lespade, Georgia Hulme | | 12 min read
Mass spectrometry imaging (MSI) simultaneously acquires increasingly detailed molecular and spatial information, opening up new worlds of research possibilities. And though this rapidly-developing discipline can be widely applied to a multitude of fields, including food, environmental, and forensic analysis, its applicability to biological samples looks set to reshape the field of clinical diagnostics.
We spoke to Eva Cuypers, Assistant Professor, University of Maastricht, the Netherlands, to find out more.
Meet Eva Cuypers…

After studying pharmacy at university, Cuypers obtained a PhD degree in biomedical sciences at the School of Pharmaceutical Sciences, KU Leuven, Belgium. She then undertook postdoctoral training in Forensic Toxicology at KU Leuven and at AMOLF, The Netherlands. In 2015, she was appointed as Assistant Professor in Forensic Toxicology at KU Leuven. Her research mainly focused on developing new methods in forensic toxicology – including developing novel drug screening methods for police forces and creating a compound mix for cadaver dog training.
Cuypers’ first time working with mass spectrometry imaging (MSI) was in Ron Heeren’s lab at AMOLF, where she investigated drug distribution and decontamination methods for hair analysis. Having discovered the endless possibilities of MSI, she moved to the Maastricht MultiModal Molecular Imaging Institute (M4i) at University Maastricht as a Senior Researcher in 2019. In 2021, she was appointed as Assistant Professor, and became the leader of her own research group – where she now focuses on using MSI in perioperative diagnostics. Cuypers and her team are working on MSI-based methods to detect biomarkers in biological samples. They are also advancing intraoperative diagnostics for “on-the-spot” identification of tissue or disease type, tumor border, and cancer cells. According to Cuypers, her main goal is to translate molecular insights into clinical solutions for personalized medicine.
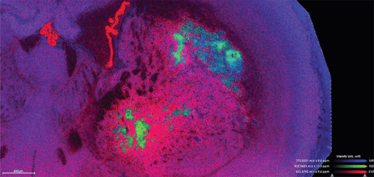
Figure 1: MALDI2-TOF image (TimsTOF, Bruker, positive mode) of rat brain showing some distinct lipid distributions in the tumor core (green) and surrounding invasive brain tissue (red). Spatial resolution: 10X10um.
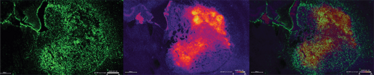
Figure 2: Left: Astrocyte IHC staining (GFAP) of rat brain tumor tissue after MSI.
Middle: MALDI2-TOF image (Positive mode, 10X10um) of the same rat brain tumor tissue showing the distribution of m/z 820.52.
Right: Overlay of astrocyte staining and the MSI image.
What inspired your interest in diagnostic mass spectrometry?
My background as a pharmacist – specializing in toxicology and analytical science – brings basic and applied sciences nicely together. My goal has always been to create technology that moves through the translational stage and into the clinical setting or forensic field – or, in other words, research that truly makes a difference! My connections with different clinical disciplines – and the brainstorming sessions with potential end-users – enriches my development as a scientist.
Could you briefly explore some of the potential applications of MSI?
How long have you got? To give you a flavor (!), one of my PhD students is using it to investigate the spatial distribution of taste compounds in mushrooms. The effect of environmental pollution on food is also a hot topic within the MSI field; in fact, at M4i, we are researching the distribution and health effects of microplastics in fish using MALDI-MSI. In forensics, we’ve used MALDI and SIMS imaging to investigate drug distribution in a singular hair (1). With MSI, a full timeline of drug use can be obtained with a singular experiment and an accuracy of hours. Alternative methods, such as GC-MS, take months to gather similar results. MSI also has further forensic possibilities in fingerprints, tissue and bone samples, fibers, and ink.
Finally, in clinical research, I’d say MSI has the potential to revolutionize the field. MSI can provide valuable information about the distribution and abundance of various compounds in biological samples that can inform the development of new diagnostic tools, treatments, and therapies. For example, MSI can be used to study the distribution of drugs in tissues to determine their efficiency and safety, which can help optimize drug delivery and improve drug effectiveness.
But it can also be used directly in disease diagnosis and prognosis through the identification of specific biomarkers or compounds that are associated with specific diseases in tissues or cells from patients. Here, the focus is often on improving the accuracy of disease diagnosis and/or enabling earlier detection of disease.
MSI is also useful in tissue engineering, where it can be used to examine the distribution of various compounds in engineered tissues – such as scaffolds or implants – to determine both their composition and quality. It is also vital for toxicology and offers a way to study the distribution of various compounds in tissues to determine their toxicity.
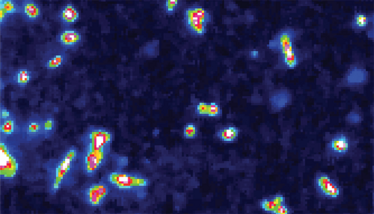

Figure 3: Breast cancer 2D cells (MDA-231) imaged with MALDI-MS, positive mode, 5x5um spatial resolution. Colors indicate the peak intensity of the selected m/z and showing a higher concentration in the cell core compared to the cell surrounding.
Could you share some of the techniques you’ve helped develop in your career?
I have developed many standardized analytical methods during my time as a forensic toxicologist, including new extraction methods for dried blood spots and the use of ionic liquids as micro-extraction solvents for blood analysis. The research papers that I am most proud of are the ones that have had an advantageous impact on the end-user field – for example, my extraction and detection method for explosives in human blood and tissue, and the development of guidelines for intervention staff dismantling illegal cannabis plantations.
My MSI-related research on the consequences of decontamination procedures in forensic hair analysis was extremely significant. The method gave fresh insight into drug incorporation in hair and changed routine protocols around the world. I’m very proud to say that I won three international awards with this research!
My latest award-winning research was on the development of an intraoperative detection and identification method for brain tumors – the start of a new intraoperative diagnostics era. Lastly, my ongoing research on single cell MSI to connect laboratory research with clinical diagnostics is one with high impact potential. I am excited to see this research progress towards clinical implementation of MSI in digital pathology.
You’ve also used MSI in breast cancer research…
That’s right. I developed a method where two-dimensional cells can grow on glass slides for MSI measurements – a technique that allowed us to generate molecular profiles from single cells with a spatial resolution of five micrometers. Alongside our collaborators, we built a molecular database and recognition system of 14 different breast cancer cell lines and successfully applied this system by correctly identifying the cell types in tissue samples.
Pathological stainings and their interpretation are often still done manually by looking through a microscope – resulting in a rather ‘subjective’ interpretation that depends on the pathologist's experience and domain of expertise. Our MSI method can indicate on-the-fly the cell type in an objective manner based on the molecular profiles. Moreover, the cell type recognition can be done ‘on-the-fly’ without the laborious and time consuming stainings. The cell identifications were even possible during the MSI run (on-the-fly) – making it a fast and objective method.
The advantage of MSI here is that it is a label-free method, and therefore not limited to the amount of histological staining possible on one tissue sample. We published this research last year but also filed a patent for “digital on-the-fly pathology” and we are currently searching for a company or private partner to invest in this.
Recognizing single cells in a tissue environment – without the need for labeling – has opened up new research possibilities. Molecular changes of cell types in tumor tissue can now be detected before microscopic cell changes are even visible, which can inform therapy outcome predictions. More specifically, we showed that clinical diagnostic and prognostic markers, such as receptors, can be linked to a molecular profile, which means we can recognize them using the same single cell molecular database.
In addition, the role of specific cell types and cell–cell interactions in tumor development and progression can be investigated with a spatially detailed method that gives in-depth knowledge and further insight into specific diseases. Undoubtedly, this molecular-based, single cell recognition strategy will lead to faster and more targeted drug development.
Notably, our proof-of-concept molecular database method can be applied to any cell type. The future goal is to build cell-specific databases for different diseases and investigate whether linked recognition systems can be used for early diagnosis and prognosis. In the next few years, my research group will mainly focus on brain tumors and neurodegenerative diseases.
In short, I believe MSI will have a huge impact in personalized clinical diagnostics, prognostics, and treatment.
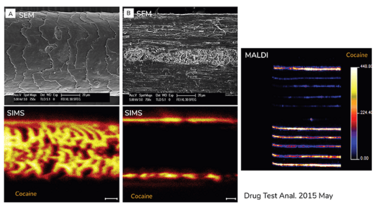
Figure4: Top: SEM images of intact (A) and longitudinal sectioned (B) hair samples.
Bottom: MetA-SIMS (Metal assisted gold coated sample, TRIFT II TOF-SIMS with Au liquid metal gun) image of cocaine distribution of intact (A) and longitudinal sectioned (B) hair that is externally contaminated with cocaine.
Right: MALDI-TOF (Synapt, 150x150um, positive mode) image of cocaine distribution on intact users hair samples.
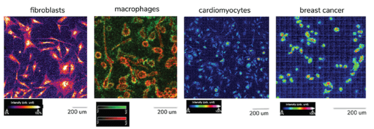
Figure 5: MALDI-TOF (TimsTOF, MALDI1) image of 2D cultured fibroblasts, macrophages, cardiomyocytes and breast cancer cells. All acquired in positive mode at 5X5um spatial resolution.
Macrophage image: two different m/z values (red: m/z 528.20, green: m/z 591,17) are selected to show the difference of their respective spatial distribution (red in core and green in cell 'outer layer’).
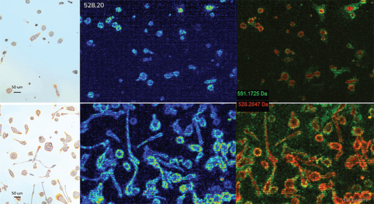
Figure 6: Left: Light microscopy images of M1 (top) and M2 (bottom) macrophages (2D cell types).
Middle: MALDI-TOF (Rapiflex, positive mode, 5x5um) image of the distribution of m/z 528.20 in respectively M1 (top) and M2 (bottom) macrophages.
Right: MALDI-TOF (Rapiflex, positive mode, 5x5um) image of the distribution of m/z 591.17 (green) and m/z 528.20 (red). Both compounds show a distinct intracellular distribution.
How will MSI techniques mesh with other emerging diagnostic trends, such as digital/computational pathology and artificial intelligence?
MSI needs to be combined with artificial intelligence (AI) to be properly applied in routine diagnostic practice. The advantage – and disadvantage – of MSI is that it generates very detailed spatial and molecular information, which means there is simply too much data for the human brain to process in an acceptable time frame. AI can teach the system to distinguish between healthy and diseased tissue – a crucial task for the introduction of MSI into point-of-care settings.
Our research on breast cancer cells has shown us that lipids are a very important group of molecules. Lipids play a major role in cellular structure, energetics, and signaling. The characterization of changes in cellular lipid composition is a key research element for diagnostic purposes. In contrast to genetic profiling, lipidomics gives greater insight into cell activity, communication, and environmental changes occurring in the tissue. Although histological stainings are useful for different cell types or necrotic and hypoxic regions, they do not give detailed information on the cell energy level or cell signaling – both crucial parameters in cancer progression. However, there is not one, singular lipid that acts as a cancer marker. Rather, it is the change in lipid ratios that specifies cell type, and these changing ratios can only be analyzed for diagnostic purposes in a reasonable time frame when machine learning is used.
Our work on single cells is a fundamental step towards the direct translation and integration of MSI into point-of-care settings, and close collaboration with pathologists and surgeons is key to our success. MSI-based diagnostics will be used alongside current pathology workflows, and provide pathologists with more detailed molecular information. Diagnostic and prognostic interpretation should be closely developed with clinicians for direct patient implication, as it is highly important to include end-users as early as possible to ensure clinician usability and potential.
How can analytical scientists and clinicians work more closely together?
At Maastricht University Medical Center and UZ Leuven – two clinical centers I am collaborating with – most clinicians are open to new research and diagnostic methods. I believe the key factors to a successful partnership are communication and scientific curiosity. And that means a willingness from both sides (clinicians and researchers) to put in time and effort. As a researcher, I take the time to observe and discuss surgery and diagnostic procedures. I identify the strengths and weaknesses of a clinician’s methods and tools because they are the most interesting parts of my research to tackle. I am always willing to adapt proof-of-concept methods depending on clinicians’ input. On the other hand, a clinician should spend time explaining procedures, testing new methods, and comparing these with existing clinical strategies.
A great example of the importance of communication can be found in our intraoperative brain tumor project. As researchers, we thought it would be advantageous to develop a method that could identify tumor type and grade. However, after conversing with the neurosurgeons, it became clear that this was an unnecessary endeavor. Instead, neurosurgeons needed a method that could accurately and sensitively detect the glioma tumor border. So, based on their input, we changed our whole recognition system. Open communication is crucial and necessary to adapt and revise methods so they can have clinical relevance in the future.
Practical logistics are also important considerations in this line of research; M4i is situated right next to the hospital, which allows daily interaction and easy sample transfer.
Are clinicians – and especially pathologists – concerned about MSI technology encroaching on their territory?
Certainly, MSI will revolutionize clinical diagnostics; however, I’m always quick to point out that it will supplement the pathologist’s job, rather than replace it. Although AI recognition methods are quickly evolving – they will always require human interpretation before decisions can be made about a patient's treatment.
What lies ahead for your work – and for MSI and lipidomics in medicine more generally?
In the past five years, MSI has evolved massively in terms of sensitivity, mass accuracy, and spatial resolution – and it is only a matter of time before the technology becomes a major part of clinical diagnostics. Single cell resolution – necessary for digital pathology – has only recently become possible. Although there are challenges – such as speed and data size – that need to be addressed, new methods are being developed by M4i to significantly speed up analysis time, while making sure single cell spatial resolution is maintained. When a sample can be scanned in a matter of seconds, a major step will be taken to adopt digital pathology worldwide. Datamining research is also moving fast and AI will be an indispensable factor.
The MSI field is continuously evolving, and the question is no longer if it will move into clinical practice, but when will it happen?
- E Cuypers et al., Anal Chem, 88, 3091 (2016). DOI: 10.1021/acs.analchem.5b03979
Associate Editor, The Cannabis Scientist
Associate Editor, The Analytical Scientist















