
DNA on the Cutting Room Floor
CRISPR technology has revolutionized genomic editing – but is there still a place for first-generation tools?
There’s no doubt CRISPR (clustered regularly interspaced short palindromic repeats) technology has been a game changer for genome editing. Cheap, easy, and efficient, it has resulted in an explosion of research in the field, with translational scientists using the new tool for everything from studying the origins of cancer to developing gene therapies that replace “faulty” genes. However, it is by no means the first or only technology to allow genome alteration. And though CRISPR may be the “latest and greatest”, it isn’t necessarily the best option for every application.
There are three main categories of engineered nucleases on the market: zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR. All are targeted “genomic scissors” that combine an engineered DNA-binding protein with a DNA-cleaving enzyme to make a double-strand break at a specified location in the organism’s DNA. After the relevant sequence has been snipped out, the breaks are sewn neatly back together by the cell’s own repair machinery.
“It is a very exciting time for people like me who are interested in genome editing and now have access to this great toolkit,” says Jon Chesnut, Senior Director of Synthetic Biology and R&D at Thermo Fisher Scientific
DNA scissor debut
ZFNs were the first onto the market and opened up a whole new world, recalls Supriya Shivakumar, Head of Strategy and Portfolio Development, Gene Editing and Novel Modalities at MilliporeSigma, which licenses the technology for research use.
“We already had the ability to downregulate genes using RNA, but we were missing a piece of the puzzle – the ability to knock out a gene entirely,” says Shivakumar.
Each zinc finger recognizes three or four bases, and by combining these modules in different configurations, almost any sequence can be targeted. ZFNs are heterodimers, with the zinc fingers joined to a FokI endonuclease domain that cleaves the DNA. FokI will only produce the desired double-strand break when dimerized, which increases specificity by ensuring that cleavage only occurs when two DNA-binding events occur together.
ZFNs cannot really be described as “do-it-yourself” technology – they are time consuming and expensive to make, so most users turn to vendors for custom-made solutions. “ZFNs are hard to get completely right. We have an archive of experimentally-derived information, and without it there is a real chance that longer nucleotide recognition sequences won’t work,” says Shivakumar.
An alternative to ZFNs became available in the late 2000s – TALENs. They harness plant pathogen proteins (TAL effectors) for DNA binding, again linked to a nuclease that snips the DNA.
TALENs are built from arrays of 33–35 amino acids – each array targets a single base pair. Constructing DNA-binding domains with TALENs is less complex than with ZFNs, meaning that labs can save money by creating their own. However, they are large molecules, which can make them difficult to deliver efficiently, and still require a level of skill to engineer.
CRISPR fever
“TALENS and ZFNs are expensive and time-consuming, and so are typically used when you want to create a specific cell line. What was missing was the ‘what if?’ – the ability to play and explore,” says Shivakumar. Enter CRISPR/Cas.
CRISPR/Cas originated as a form of adaptive immunity in bacteria. When a bacterial cell is infected, short sequences of the viral genome (CRISPRs) are incorporated into the bacterial genome. CRISPR-associated (Cas) proteins process these sequences and attack the virus by cutting matching DNA sequences in the viral genome. By engineering plasmids encoding both CRISPRs and Cas, a hugely efficient genomic editing tool is created.
“The biggest benefit to CRISPR/Cas is ease of design,” says Chesnut. “You can change the specificity of the nuclease just by changing the sequence of a small piece of RNA, which has really opened up genome editing.”
Most labs are very comfortable with the technology; as Shivakumar puts it, “You can make CRISPRs in your garage.” High speed, low cost, and impressive efficacy make CRISPR suitable for screening studies. For example, researchers can knock out thousands of different genes in cancer cell cultures to find out which are responsible for drug resistance. “That puts it in a different category to ZFNs and TALENs – you can not only study known targets, but find entirely new targets,” says Shivakumar.
Precarious Patents
The well-publicized patent dispute over CRISPR/Cas looks set to run and run
Although the sequences were first described by Osaka University researchers in the late 1980s, the term CRISPR wasn’t coined until the early 2000s. The potential of CRISPR/Cas in genome editing became clear when University of California, Berkeley (UCB) researcher Jennifer Doudna and collaborators published work in 2012 showing that they had reprogrammed the system to target specific sites in bacterial DNA. In early 2013, several groups, including one led by the Broad Institute’s Feng Zhang, reported that the system also worked in eukaryotic cells, including human cells. Both UCB and Broad Institute have filed patents covering the fundamentals of genome editing by CRISPR/Cas, and licensed them to a number of companies. Now they are locked in a fierce dispute through the US Patent and Trademark Office (USPTO) and European Patent Office.
It’s thought the proceedings may drag on for years as the authorities try to determine whether the use of CRISPR/Cas in eukaryotic cells was an obvious follow-on from Doudna’s original work, or a separate (and patentable) breakthrough. The battle has become less diplomatic in recent months, with harsh words and accusations of impropriety being exchanged by the opposing institutions. Given the huge potential earnings for the institutions and their licensees, it seems likely that the fight will be a long and drawn-out one.
The mudslinging and high profile of the scientists involved has ensured that most of the attention has been focused on the foundational patents described above. However, a number of smaller groups have also filed important patents relating to CRISPR/Cas, including MilliporeSigma. “It’s interesting that the whole focus has been on the dispute between these two major scientific institutes. I think there are some dark horses in this race,” says MilliporeSigma’s Supriya Shivakumar.
Many companies now provide off-the-shelf or custom-designed libraries for screening studies, says Chesnut. “We just launched our Lenti-based arrayed CRISPR libraries, and we hope to cover the entire human genome by early 2017.”
Shivakumar also notes the rise of library platforms: “We don’t want to convince someone to buy CRISPRs from us if it is quicker and cheaper to do it themselves, so we haven’t focused on making individual CRISPRs, but rather making whole-genome libraries with the Sanger–Wellcome Trust and the Broad Institute.”
Ultimately, what makes CRISPR so exciting is the freedom it offers. “The ‘sandbox’ environment that CRISPR enables is where great leaps in science originate,” says Shivakumar. “It has opened the door to a number of new technologies that we never predicted.”
Cut to the chase
If CRISPR is the fastest, cheapest, and most efficient technology, why would you consider using ZFNs or TALENs? Well, there are some situations in which the older technologies actually work better, says Chesnut. “We know of applications where TALENs are technically more effective than CRISPR. In transcriptional modification, TAL effectors appear to be at least as efficient as CRISPRs in delivering transcriptional activators and repressors.”
Particularly relevant to the translational field is the fact that CRISPR/Cas is still very much the new kid on the block. “With CRISPR, you can play. But once you have a target that works, there are a couple of reasons to switch to a more established tool, such as ZFNs or TALENs,” says Shivakumar. “ZFNs were one of the first technologies on the market and there has been a lot of work to show that there are no off-target effects or toxicity. It was one of the first to be used clinically, so there is a proven pathway.”
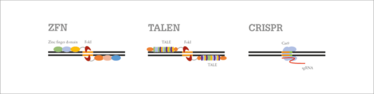
Figure 1. Mechanisms of ZFN, TALEN and CRISPR/Cas9 genome editing.
The IP situation surrounding the newer technology could also prove a major headache for those looking to bring a CRISPR/Cas-based gene therapy to market. The ongoing patent dispute (outlined in “Precarious Patents”) could be a source of unwelcome uncertainty for years to come. “We are watching the patent dispute around CRISPR closely, both in terms of how it may affect our customers, and as a fascinating view into how an emerging technology can stir up so much discussion. Scientists aren’t usually so emotionally invested in the subject – at least in public,” says Shivakumar.
“There’s a significant amount of confusion about who’s going to end up owning the IP rights for CRISPR, in contrast to the clear IP landscape for TALENs and ZFNs,” says Chesnut. “It comes down to what your end goals are. If you want to create SNP-edited or knockout cell lines, certainly CRISPR seems to be the go-to. But if you are thinking further ahead and want to commercialize, then you have to consider IP.”
Gene editing: the sequel
Will CRISPRs be all-conquering, or will they themselves be replaced by the next big breakthrough technology? What great advances in science and medicine will result? The field is moving so fast that it’s hard to tell what the future holds. “Whatever happens – it’s going to be surprising,” says Shivakumar.
Gene therapy – a dirty word just a decade ago – is once again a major research focus, bolstered by the success of CAR-T cell therapy for cancer. “In our lifetime, I think we will see gene and cell therapy in regular use,” says Shivakumar.
Genome editing is also finding applications in the pharmaceutical industry, creating in vitro models for drug safety and toxicity testing, and replacing RNAi screens for discovery. “We devote a lot of effort to working with pharma customers to use genome editing to create better cell screening models for drug discovery – both specifically edited cell lines and screening library approaches,” says Chesnut.
Shivakumar believes that using CRISPR in detection technologies could have a big impact on day-to-day life. For example, farmers could grow plants that would change color to indicate nitrogen levels in the soil. In medicine, this could take the form of sophisticated sensors for disease detection and monitoring. “Perhaps one day we’ll all have the equivalent of a Star Trek tricorder,” she suggests.
However, lest we get swept away by the brave new world of CRISPR/Cas, Shivakumar also strikes a note of caution. “With the explosion of papers coming out, there has been a sense that if you tie your work to CRISPR, it will get a wider audience. So my advice to researchers new to the field would be to regard reported advances with a critical eye, and make sure you are using the right assay for your needs. No matter what you are doing – the better your assay, the better the process.”

“As Editor of The Translational Scientist, I’m working closely with our audience to create vibrant, engaging content that reflects the hard work and passion that goes into bringing new medicines to market. I got my start in biomedical publishing as a commissioning editor for healthcare journals and have spent my career covering everything from early-stage research to clinical medicine, so I know my way around. And I can’t think of a more interesting, challenging or important area to be working in.”















