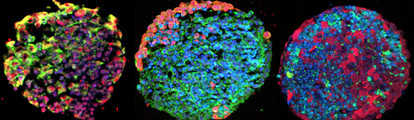
The Story and Science Behind the "Mini-brain"
How iPSC-derived 3D organoids and high-throughput histology opens a new door to personalized medicine development and testing.
A few years ago, we had successfully developed technology for high-throughput histology of 3D organoids. Meanwhile, Thomas Hartung at John Hopkins University had been working on "mini-brain" technology derived from iPS cells. After initial discussions at a meeting of the Society of Toxicology in 2016, we quickly recognized that working together could be a great opporunity. We could better demonstrate the utility of our technology, while Thomas could further explore the potential of the mini-brain model.
What came up again and again in our discussions was the need for new therapies for brain cancer. One of the reasons there has been little progress in the field was because of a lack of proper preclinical drug testing models; both for brain tumors and indeed for many brain diseases, including Alzheimer’s and Parkinson’s. Stem cell technologies offer hope in that regard because it’s now possible to build human-relevant 3D models. Many of these models – derived from human neuronal progenitor cells – seem to replicate some aspects of brain functions. However, working with 3D tissue models as a means to screen and test drugs is challenging. The main reason? A lack of robust technologies for multiplex analysis. Many of the technologies available exist in whole mount format, using confocal microscopy. There are general challenges imaging whole spheroids in 3D – light scatter and antibody penetration being two – but multiplex analysis brings additional challenges in terms of staining, which often involves multiple steps of antigen retrieval, potentially damaging the organoids and compromising the data.
We developed our technology to overcome these challenges. By layering the microtissues into a microarray format, replicating multiwell plates, it is possible to image tissue without resolution or light scatter issues. Very thin sections are cut through all the spheroids, which facilitates standard staining technologies and standard confocal microscopy. It’s also more practical from a data-crunching point of view – the dataset generated from these 96 well plates is somewhat smaller than through optical imaging in 3D.
In the paper we’ve just published (1), we test a variety of drugs against glioblastoma using the model – an approach that resulted from our discussions with Thomas. We focused on glioblastoma because of the lack of any real progress in terms of treatment. We showed that we were able to produce a mini-brain model containing neuronal cells and other brain components – glial cells, astrocytes, oligodendrocytes, together with a glioblastoma tumor. Such a model gives us a great opportunity to assess the impact of drugs on both cancerous and normal tissue. In my opinion, being able to generate a pre-clinical model for this purpose is a real breakthrough.
There are many human tumor models currently under development, focusing on many different components of tumor biology. Models that focus on the stromal component of tumors and the immune-modulatory component that exists in tumors are particularly important, because such studies are difficult to conduct in animals. It has been tried in orthotypic and syngeneic mouse models, but the reality is that you are, of course, working with a mouse immune system rather than a human’s. With human tumor models, cells can be derived from specific sources (there are very large banks of patient-derived xenograft tumor cells, for example), which opens up the potential of personalized medicine. You could even test different drugs for a given patient’s tumor.
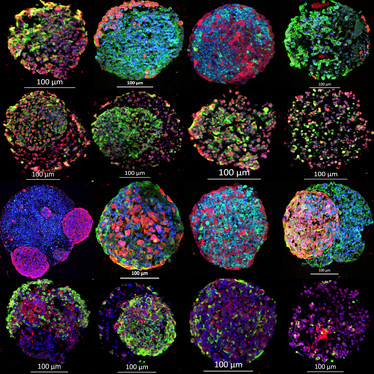
Figure 1 - Multiplex immunofluorescence stained gBS spheroid sections in the microTMA.
Red and green fluorescence represents a range of different biomarkers for both normal neuronal cells and (neurons, glial cells, astrocytes, oligodendrocytes) and glioblastoma tumor cells in the gBS. The blue colour represents a nuclear counterstain (DAPI).
Such models are also relevant to chemical safety assessments – consider the long-term tests for carcinogenicity, neuro-toxicity and developmental-toxicity. These assessments are very expensive and not always useful or relevant because they are conducted in rodent models. Human tissue models like the mini-brain could be used to better assess the risk – or at least the risk relevance.
Admittedly, there is still some way to go. Adopting 3D human tissue models in more conventional drug testing regimes will require significant validation effort. Such work could entail testing the models’ ability to predict the efficacy of a battery of known (clinically-used) drugs. Indeed, we’re working with groups from around the world to validate different disease models, and to develop new disease models. As that work is completed, I think we will see wider use of these approaches – and in a more standardized format.
- S Plummer et al., “A Human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine,” Sci Rep, Published online. (2019) PMID: 30723234.

Managing Director of Micromatrices, Dundee, UK















