Understanding Pancreatic Cancer Prognosis
Mutations in four main genes influence survival rates of pancreatic adenocarcinoma patients
Despite increased research efforts in recent years, survival rates for pancreatic cancer remain relatively low, with only 3.3 percent of adults surviving for five years with the disease (1), and researchers around the globe are working hard on new directions to aid in diagnosis, prognosis and treatment.
In a recent publication in JAMA Oncology, a group of US researchers studied 356 patients with resected pancreatic adenocarcinoma, and identified changes in four main driver genes that were associated with outcomes following surgery. Protein expression and DNA alterations for KRAS, CDKN2A, SMAD4, and TP53 were analyzed using immunohistochemistry and next generation sequencing. The research team found that patients with KRAS mutant tumors had worse disease-free and overall survival than patients with KRAS wild-type tumors. In particular, patients with KRAS G12D mutations had poorer outcomes, with a median survival of 19.7 months (2). The number of mutations present also influenced survival (see Table 1).
The authors hope that a better understanding of the molecular changes affecting patient outcomes could improve treatment approaches, and two of the collaborators – David Linehan and Brian Wolpin – are partnering on a further project investigating new therapies and biomarkers for use in metastatic pancreatic cancer (3).
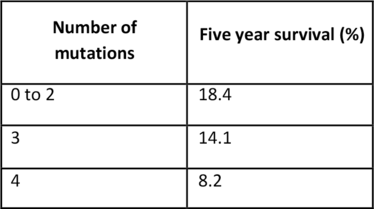
Table 1. Number of altered genes versus five year survival.
- Cancer Research UK, “One-, five- and ten-year survival for pancreatic cancer”, (2014). Available at: bit.ly/21d4tyO. Accessed November 13, 2017.
- ZQ Qian et al., “Association of alterations in main driver genes with outcomes of patients with resected pancreatic ductal adenocarcinoma”, JAMA Oncol, [Epub ahead of print] (2017). PMID: 29098284.
- Eurekalert, “Scientists link pancreatic cancer survival to four genes”, (2017). Available at: bit.ly/2zyKAwn. Accessed November 13, 2017.

I have an extensive academic background in the life sciences, having studied forensic biology and human medical genetics in my time at Strathclyde and Glasgow Universities. My research, data presentation and bioinformatics skills plus my ‘wet lab’ experience have been a superb grounding for my role as a deputy editor at Texere Publishing. The job allows me to utilize my hard-learned academic skills and experience in my current position within an exciting and contemporary publishing company.















