
State-of-the-Heart Tissue Models
A decade after Shinya Yamanaka brought us induced pluripotent stem cells, our ability to generate an endless supply of human heart cells is being put to good use in research and development.
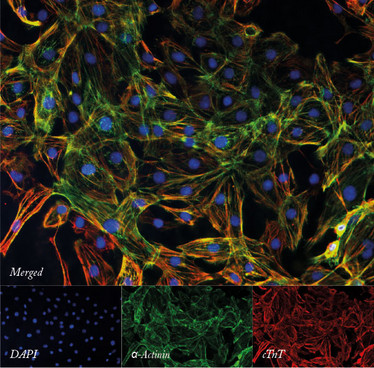
The quality and purity of cardiomyocytes differentiated from induced pluripotent stem cells can be assessed using immunofluorescence staining for cardiac-specific markers.
It is estimated that only eight percent of new drugs will successfully transition from clinical trials to market launch. Even when drugs do gain regulatory approval, some are still pulled from the market due to unforeseen complications, with cardiovascular complications often cited as the most common reason for these withdrawals (1, 2). Given our current inability to effectively model how drugs affect the heart, it is not surprising that the decreasing rate of drug development has hit cardiovascular therapies especially hard. Between 2000 and 2009, the number of new cardiovascular drugs approved by the FDA decreased by 33 percent compared with the preceding decade (3). This is particularly concerning given that, according to the American Heart Association, cardiovascular disease is already responsible for one in every four deaths in the United States and by 2030 will kill 24 million people per year worldwide (4). Despite the urgent need for better cardiovascular drugs, the lack of cardiac models capable of accurately reflecting human physiology has led the FDA to place stringent demands on cardiovascular trials, thereby further increasing their size, duration, and cost. Thus, there is a strong need for cardiac models with more accurate prognostic capabilities to enhance general drug toxicity screening and novel drug development.
Current models
While it is possible to isolate adult heart cells from patients after heart surgery, the difficulty of obtaining such tissue is further compounded by the fact that human cardiomyocytes rarely divide and do not survive more than a week when grown on a petri dish, severely limiting their application for larger-scale studies. In lieu of primary heart cells, screens for cardiotoxicity and investigations of novel drugs rely on different in vitro and in vivo models. Unfortunately, current models are either overly simplistic or not indicative of human heart function. Two of the most common in vitro models are transgenic Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK293) cells that overexpress human cardiac ion channels (5). While easy to culture and quick to divide, these heterologous cells cannot accurately emulate the complex multichannel relationships that direct the function of a real human beating heart cell. In vivo mouse models are also common and represent a more dynamic environment to study cardiovascular systems. However, mice have vastly different physiology compared to humans. For instance, on average a mouse heart contracts roughly seven times faster than a human heart (6). These deficiencies ultimately mean that many drugs fail in human trials when cardiac safety cannot be adequately established using conventional models; and conversely, some drugs with a significant risk of inducing serious cardiac complications – including seizures and sudden cardiac death – may slip through the safety net (7).
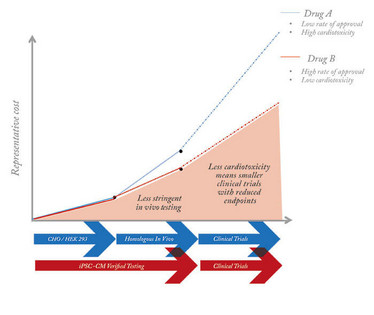
Figure 1. The current drug discovery process is limited by the cardiac models used, which increase costs and restrict development.
A new model for research
The emergence of human induced pluripotent stem cells (hiPSCs) presents the opportunity to redesign the current drug development paradigm – and to avoid some of its problems. hiPSCs were first generated in 2007 by the overexpression of four transcription factors that revert adult differentiated cells to an embryonic stem cell-like state (8). Similar to embryonic stem cells, hiPSCs have the capacity to differentiate into any cell type, and ever-more efficient derivation protocols now provide ready access to a limitless supply of human heart cells (hiPSC-CMs) (9). In addition, hiPSC-CMs retain the genetic mutations of individual donors, a fact that allows researchers to investigate the potential for individualized therapies (7, 10). Two types of cardiac diseases commonly studied using hiPSC-CMs are cardiomyopathies, which involve the deterioration of heart muscle contraction, and channelopathies, which involve the repolarization of ion channels. So far, hiPSC-CMs have been used to model several different cardiomyopathies, such as dilated, hypertrophic, and arrhythmogenic right ventricular cardiomyopathy (11-13). The most commonly studied channelopathies are those that induce long QT syndrome, a prolonged action potential duration that can cause arrhythmias, seizures, and sudden death (10, 14, 15). Preliminary channelopathy studies not only found that disease phenotypes were expressed in hiPSC-CMs, including prolonged action potential and reduced repolarization velocity, but also demonstrated their physiological response to beta blockers, a common class of cardiovascular drugs that slows the heart rate. Further, by correlating how the efficacy of different beta blockers can vary based on patient genetics, studies have demonstrated the potential of hiPSC-CMs for personalized medicine.
However, a major limitation for the use of hiPSC-CMs for drug discovery and toxicity screening is the relative immaturity of the cells compared with cardiomyocytes in adult primary heart tissue. There are many differences in genetic and phenotypic properties, including cell size, sarcomere organization, calcium handling dynamics, cardiac-specific genetic expression, and others (16). To bridge this maturation gap and improve the predictive ability of hiPSC-CMs, engineered constructs have recently been developed to culture cells in a more natural setting that recreates the physical and topographical cues of the heart. As hiPSC-CMs mature, the relevant structural and functional abnormalities caused by mutations are more easily identified. One identification metric is the alignment and uniformity of cellular sarcomeres. Similar to the coils within a spring, the sarcomeres within cardiomyocytes are the mechanisms of contraction. However, the hiPSC-CMs cultured in traditional monolayers in plastic petri dishes are often so disorganized that there is little room to distinguish between healthy and diseased cells. To address this issue, researchers at Harvard used micropatterned substrates to culture hiPSC-CMs with a mitochondrial cardiomyopathy, which increased the average sarcomere alignment, improving the discrimination of healthy and diseased cells (17).
Other engineered constructs like collagen-based engineered heart tissue allow us to directly analyze contraction, instead of relying on indirect indicators of contractile force such as calcium handling and sarcomere alignment. Engineered heart tissue is cultured on composite pillars that are stiff enough to apply passive stress but also flexible enough to measure contractile forces by the length of their bending, mimicking the tension within the natural heart. A recent study used engineered heart tissues to model dilated cardiomyopathy, identifying a mutation in a sarcomere protein that causes a decrease in contractile strength (18). Crucially, this phenotype was not otherwise evident in single-cell assays, highlighting the importance of tissue-scale disease models. In addition, multiple studies have illustrated that effective doses of beta blockers are closer to physiologic levels when administered to 3D cardiac constructs compared to traditional 2D cell culture systems (19). Taken together, these studies showcase how applied tissue engineering is improving cardiac models, supporting the use of hiPSC-CMs as potential clinical diagnostic tools that can reflect in vivo human physiology.
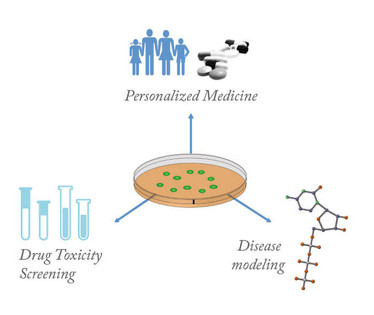
Figure 2. Applications for cardiomyocytes from induced pluripotent stem cells.
A new model for drug screening
While improved maturation can help increase the accuracy of cardiac disease models, progress is also being made in scaling these constructs for use in drug development – specifically, to enable high-throughput screening for cardiotoxicity. To this end, the field is tailoring in vitro platforms toward more automated processes. For instance, the use of microelectrode arrays allows us to investigate electrophysiology as a functional readout for drug efficacy without killing the cells. Recently, a microelectrode array was used to screen for drug-induced arrhythmias in hiPSC-CMs and to accurately identify cardiotoxic drugs (20). In addition, microfluidic cardiac chips reduce culture costs while enabling the physiological administration of drugs in a continuously circulating flow. These microfluidic chips are often paired with bioprinting, which enables researchers to consistently create spatially organized tissues with microscale resolution. Continued development of these automated, scaled systems will further cut costs, streamline analysis, and provide an unbiased platform to screen a wide range of drugs simultaneously. hiPSC technology offers clear potential to improve the drug development process by increasing the meaningful identification of novel targets while streamlining the process of cardiotoxicity screening. The development of more mature, scalable constructs will likely promote the commercial application of hiPSC-CMs to a wider range of diseases. In the future, these hiPSC-CMs may help make drug development less costly and therefore lower the barrier of entry for new cardiovascular therapies.
- J Lexchin, "How safe are new drugs? Market withdrawal of drugs approved in Canada between 1990 and 2009", Open Med. 8, e14–9 (2014).
- R McNaughtonet al., "An investigation into drug products withdrawn from the EU market between 2002 and 2011 for safety reasons and the evidence used to support the decision-making", BMJ Open 4, e004221 (2014).
- CB Fordyce et al., "Cardiovascular drug development: is it dead or just hibernating?", J Am Coll Cardiol 65, 1567–1582 (2015).
- AS Go et al., "Heart disease and stroke statistics--2014 update: a report from the American Heart Association", Circulation 129, e28–e292 (2014).
- E Matsa et al., "Human stem cells for modeling heart disease and for drug discovery", Sci Transl Med 6, 239ps6 (2014).
- J Rice, "Animal models: Not close enough", Nature 484, S9–S9 (2012).
- SR Braam et al., "Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery", Trends Pharmacol Sci 30, 536–545 (2009).
- K Takahashi et al., "Induction of pluripotent stem cells from adult human fibroblasts by defined factors", Cell 131, 861–872 (2007).
- PW Burridge et al., "Chemically defined generation of human cardiomyocytes", Nat Methods 11, 855–860 (2014).
- I Itzhaki et al., "Modelling the long QT syndrome with induced pluripotent stem cells", Nature, 471, 225–229 (2011).
- N Sun et al., "Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy", Sci Transl Med 4, 130ra47 (2012).
- C Kim et al., "Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs", Nature 494, 105–110 (2013).
- F Lan et al., "Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells", Cell Stem Cell 12, 101–113 (2013).
- A Moretti et al., "Patient-specific induced pluripotent stem-cell models for long-QT syndrome", N Engl J Med 363, 1397–1409 (2010).
- E Matsa et al., "Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation", Eur Heart J 32, 952–962 (2011).
- E Tzatzalos et al., "Engineered heart tissues and induced pluripotent stem cells: Macro- and microstructures for disease modeling, drug screening, and translational studies", Adv Drug Deliv Rev 96, 234–244 (2015).
- G Wang et al., "Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies", Nat Med 20, 616–623 (2014).
- JT Hinson et al., "Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy", Science 349, 982–986 (2015).
- A Mathur et al., "Human iPSC-based cardiac microphysiological system for drug screening applications", Sci Rep 5, 8883 (2015).
- EG Navarrete et al., "Screening drug-induced arrhythmia ssing human induced pluripotent stem cell–derived cardiomyocytes and low-impedance microelectrode arrays", Circulation 128, S3–S13 (2013).
John Ahrens is a recent graduate of Stanford University with a degree in Biomechanical Engineering. His research has focused in the application of induced pluripotent stem cells as models for drug discovery and disease modelling.
Joseph C. Wu is Director of the Stanford Cardiovascular Institute and Professor in the Department of Medicine (Cardiology) and Radiology at the Stanford University School of Medicine. His lab works on biological mechanisms of patient-specific and disease-specific induced pluripotent stem cells (iPSCs). The main goals are to (i) understand basic cardiovascular disease mechanisms, (ii) accelerate drug discovery and screening, and (iii) develop personalized medicine and “clinical trial in a dish” platforms. His lab uses a combination of genomics, stem cells, cellular & molecular biology, physiological testing, and molecular imaging technologies to better understand molecular and pathophysiological processes. His clinical interests as a cardiologist involve cardiovascular imaging and adult congenital heart disease.















