Metabolomics: the Superglue of Omics
As the profile of metabolomics soars, we offer our thoughts on the future of the field.
Our understanding of cellular and molecular processes has increased exponentially during the last century. Several key questions about the function of our cells and, to a broader extent, their interactions with entire organisms have been answered, with the most prominent findings being the elucidation of our DNA structure and the sequencing of the human genome (1). The key outcome of 20th century biomedical and biological research was the knowledge we gained about the coding, content and flow of information within our cells and body. The concept that our cells contain our hereditary information in the form of DNA, which is transcribed into RNA and translated into proteins that are ultimately involved in the modulation of our metabolome is the central dogma of biology, and a comprehensive understanding of this process represents the fundamental goal of omics technologies (2).
But beyond a better understanding of ourselves, what has been the impact of these findings and technologies on our lives? Genomics (DNA sequencing) has certainly enhanced diagnosis and treatment of inborn errors and hereditary diseases. Transcriptomics (the analysis of RNA transcripts) complements genomics by allowing us to decipher gene expression, again helping to identify undiagnosed cases of genetic diseases. Proteomics (the comprehensive analysis of an organism’s proteins) would ideally complement genomics and transcriptomics; however, several ongoing analytical challenges are still hampering comprehensive protein analysis. Nevertheless, we believe advances in proteomics happening today will allow for a more efficient use of proteomics tomorrow, so that it may become a complementary diagnostic approach to other omics techniques (3).
Metabolomics rules
Ultimately, it is the metabolome – the underlying biochemical layer of the genome, transcriptome and proteome – that reflects all the information expressed and modulated by all other omic layers. The metabolome is the closest link to the phenotype and hence at the forefront of personalized diagnosis and therapy (4)(5). This concept is now widely accepted and has enhanced the application of metabolomics in biomarker research, but it has also blurred our view of its broader implications. A sea change is taking place, which defines the metabolome not only as an organism’s phenotype, but also includes the biological activity of metabolites and their respective control functions (6)(7)(8).
When taking a closer look at the involvement of metabolites in gene expression, transcription and translation control, it becomes evident that the metabolome is not simply a cellular information sink. Rather, the metabolome is the omics superglue, providing biochemical feedback across all omic layers (see Figure 1). Metabolites are essential in the control of gene expression, with several transcription factors controlled by specific molecules, including the liver X receptors, estrogen receptors and thyroid hormone receptors. In addition, as metabolic control has become better recognized (9), numerous other cellular receptors activated by metabolites have been discovered, including G-protein coupled receptors that are involved in signal transduction. Beyond these noncovalent protein–metabolite interactions, metabolites are also active substrates and products of enzymatic reactions, as well as participants in protein post-translational modifications, such as acetylation and palmitoylation. This activity even extends to metabolites shaping the metabolic environment to influence protein function, as illustrated by membrane proteins (10).
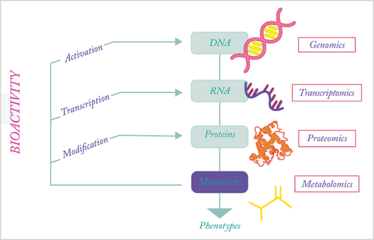
Figure 1. Metabolic activity among the different omic layers.
Clearly, the metabolome interacts with, and hence coalesces, all the omic layers. Therefore, it is not surprising that scientists in the field have started to emphasize the biological activity of metabolites as an integral part of metabolomic studies (6)(8)(11). When systems biology evolved as a biological discipline, with the express purpose of comprehensively describing all the biochemical reactions of an organism, metabolomics was rapidly recognized as an important component, allowing us to confirm computational predictions and shed light on biochemical pathways (12). However, it is only recently that the concept of metabolite activity screening (MAS) has started to integrate metabolomics, systems biology, and bioactivity (7). The field is in its infancy, yet the practical impact of identifying novel bioactive metabolites and characterizing their associated control, function, and mechanisms of action is exciting.
Critical MAS
Omega-3 poly-unsaturated fatty acids (n-3-PUFAs) and their corresponding metabolic products exemplify the power of directly connecting metabolites with health. Evolving from the observation that Inuit populations – with their n-3-PUFA rich diets – have a lower incidence of heart disease, today, n-3-PUFAs can be considered one of the most important and scientifically proven dietary supplements, with numerous beneficial health effects (13). Charles Serhan’s group at Harvard University has subsequently elucidated how n-3-PUFAs and their downstream metabolites function as immunologically active molecules, founding the field of resolution physiology and opening up novel strategies for the treatment of (chronic) inflammation (14). While the n-3-PUFA field originally sprung from a 1970s epidemiological study and the comparison of dietary habits, it is now at the cutting edge of metabolomics.
The combination of systems biology and MAS (systems-MAS), will allow us to systematically elucidate novel pathways and key metabolites, which might subsequently be used as novel therapeutics. An example is the recent systems-MAS finding that taurine significantly enhances drug-induced oligodendrocyte precursor cell differentiation and facilitates the in vitro myelination of co-cultured axons (15). This is especially interesting because metabolites are generally safe, inexpensive, readily available and can rapidly impact a system, be it a cell or an entire organism.
As with any new scientific endeavor, systems-MAS needs to overcome several significant challenges, including the need to develop effective approaches to identify active metabolites and generate libraries cataloging their bioactivity. This process is challenging because metabolic activity will likely vary depending on the organism and the phenotype of that organism. The hope is that when enough biochemical pathway and biological activity data have been correlated, we will be able to shape the metabolic landscape to our own ends. It’s possible we could even harness metabolism by stimulating the other omic layers, allowing us to better understand and manipulate physiology, to develop new therapeutics or stimulate or inhibit bacteria (influencing the microbiome). We find this concept particularly intriguing as it transforms our view of the metabolome, which changes from a phenotypic descriptor into a phenotype modulator (7). Perhaps, following on from the success of therapeutic antibodies, metabolites may evolve as a new means to “fix biology with biology”.
Martin Giera is Associate Professor and Head of the Metabolomics group at the Center for Proteomics and Metabolomics, Leiden University Medical Center, Leiden, The Netherlands.
Mary E. Spilker is Associate Research Fellow at Pfizer Worldwide Research and Development and is currently a visiting scientist at the Scripps Center for Metabolomics at the Scripps Research Institute, La Jolla, California, USA.
Gary Siuzdak is Professor and Director of the Scripps Center for Metabolomics at the Scripps Research Institute, La Jolla, California, USA.
- International Human Genome Sequencing Consortium, “Initial sequencing and analysis of the human genome”, Nature, 409, 860 (2001).
- F Crick, “Central dogma of molecular biology”, Nature, 227, 561 (1970).
- PE Geyer et al., “Revisiting biomarker discovery by plasma proteomics”, Mol Syst Biol, 13, 942–957 (2017).
- M Jacob et al., “Metabolomics toward personalized medicine”, Mass Spec Rev, 2017, 1–18 (2017).
- I Kohler et al., “Analytical pitfalls and challenges in clinical metabolomics”, Bioanalysis, 8, 1509–1532 (2016).
- T Huan et al., “Systems biology guided by XCMS Online metabolomics, Nature Methods, 14, 461 (2017).
- C Guijas et al., “Metabolites that modulate phenotype can be identified by metabolomics activity screening”, Nat Biotechnol (2018) (Accepted manuscript).
- M Giera, F Branco dos Santos, G Siuzdak, “Metabolite-induced protein expression guided by metabolomics and systems biology”, Cell Metab (2018) (Accepted manuscript).
- AS Husted et al., “GPCR-mediated signaling of metabolites”, Cell Metab, 25, 777–796 (2017).
- DA Los, N Murata, “Regulation of enzymatic activity and gene expression by membrane fluidity”, Science’s STKE, 2000, pe1 (2000).
- F Branco dos Santos et al., “Probing the genome-scale metabolic landscape of Bordetella pertussis, the causative agent of whooping cough”, Appl Environ Microbiol, 83, e01528-17 (2017).
- DB Kell, “Metabolomics and systems biology: making sense of the soup”, Curr Opin Microbiol, 7, 296–307 (2004).
- JH O’Keefe, Jr, WS Harris, “From Inuit to implementation: omega-3 fatty acids come of age”, Mayo Clinic Proc, 75 607–614 (2000).
- CN Serhan, “Pro-resolving lipid mediators are leads for resolution physiology”, Nature, 510, 92 (2014).
- BA Beyer et al., “Metabolomics-based discovery of a metabolite that enhances oligodendrocyte maturation”, Nat Chem Biol, 14, 22–28 (2018).
Associate Professor and Head of the Metabolomics group at the Center for Proteomics and Metabolomics, Leiden University Medical Center, Leiden, The Netherlands.
Associate Research Fellow at Pfizer Worldwide Research and Development and is currently a visiting scientist at the Scripps Center for Metabolomics at the Scripps Research Institute, La Jolla, California, USA.
Professor and Director of the Scripps Center for Metabolomics at the Scripps Research Institute, La Jolla, California, USA.